The COVID19 application was developed as part of the COVID19 testing initiative at the Vienna BioCenter in spring 2020. Johannes Zuber and the COVID19 testing team have established and validated a simple, safe and well-tolerable gargling procedure that can be performed at home, thereby eliminating the logistical burden and infection risk associated with centralized sampling.
Beside of the sampling procedure a web-based database solution was developed by us. We have used our SISY (Sample Information SYstem) framework for this project as it has very dynamic data structure for sample tracking systems and it is well proven through our projects, eg. iMOBS, SCCF LIMS, CRYOS and HaploidDB.
This site provides …
- an insight into our work during the design and development of the COVID19 application;
- an overview of the workflow of the COVID19 testing procedure and related features of the COVID19 application;
- more technical details of the COVID19 application and
- contact information
Development and project management
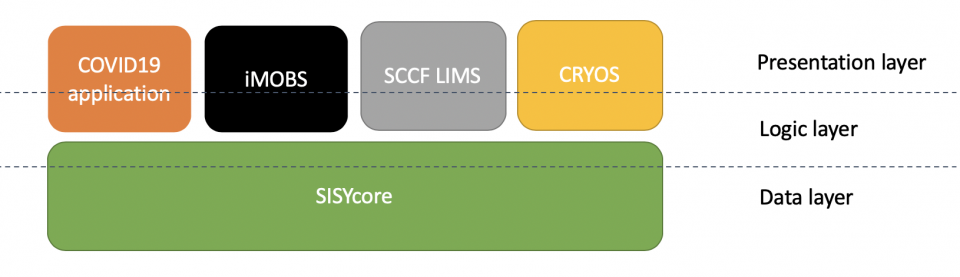
The COVID19 application is based on the SISY (Sample Information SYstem) framework which has been developed and used by other sample tracking solutions. The following figure describes the current SISY architecture:
iMOBS | integrative Management of Biological Samples
SCCF LIMS | Stem Cell Core Facility Lab Information System
CRYOS | Cryopreservation information system
SCRUM has been used as project management approach with following settings:
- Sprint length: flexible (10 – 21 days)
- SCRUM management tool: Azure DevOps
- Working item types: User stories, Bugs
- SCRUM team and roles
- Thomas Micheler (SCRUM Master & Product Owner)
- Simon Strobl (SCRUM Team & Service Owner)
- Selina Brinnich (SCRUM Team & Service Co-Owner)
- Thomas Fellner (SCRUM Team)
Testing pipeline and application features
- Patient consent form
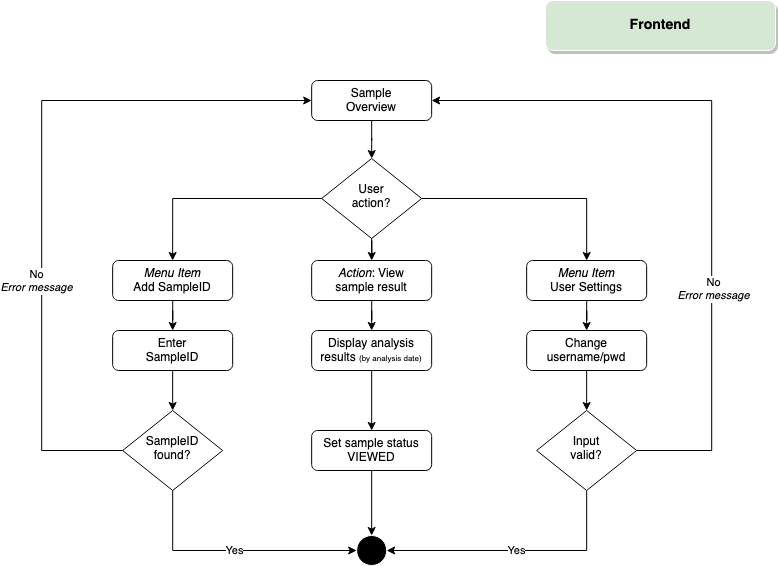
To participate in COVID-19 testing, all users fill a general consent form (detailing sample and data handling, as well as procedures in case of a positive test result). Upon declaring their consent, users are provided with an anonymous User ID/Password combination that provides access to the database frontend.
The patient consent form can be printed by the COVID19 backend team.
- (Anonymous) UserID
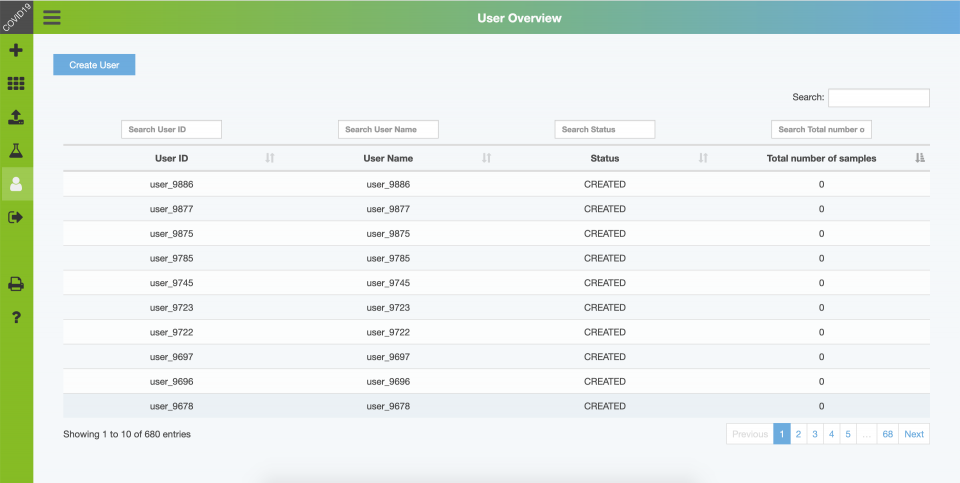
Voluntariness and anonymity is one of the major non-functional goals of this project. We have implemented a feature to create nonpersonal user accounts with an userID, password and username. Optionally more attributes can be assigned to an user account.
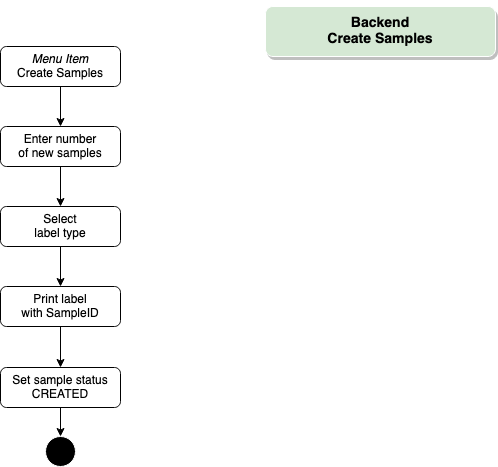
- SampleID
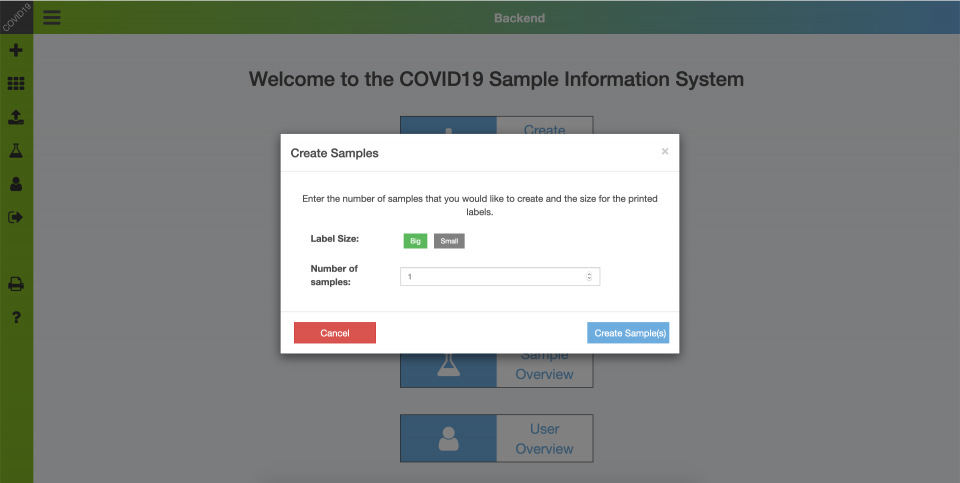
For testing, users obtain a testing kit that contains a sample tube, which is labeled with a 10- or 8-digit sample number and a code (barcode/QR). The systems supports sample tubes provided by different manufacturers. In addition, there is the option print labels and codes via the COVID19 Application. Users must note the sample number (e.g. using a photo) for result retrieval.



- QR code
The printing feature of the COVID10 application prints the sampleID on labels as human readable text and QR code which is used for further processing within the testing pipeline.

- Label, QR code printer and QR code scanner
The labels used for the COVID19 application are designed to withstand extended storage in liquid or vapor phase liquid nitrogen and ultra-low temperature freezers, for use in scientific research, clinical laboratories, and biobanks. The application supports a wide variety of containers, like cryo vials and tubes, microplates, PCR tubes, and other commonly used laboratory products.
Currently the COVID19 application supports following printer types:


- Plate administration
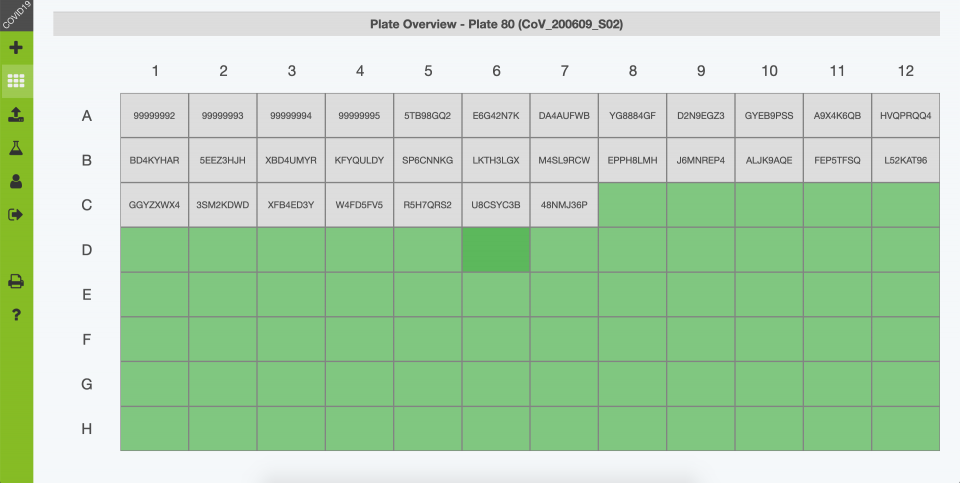
After sample submission, all collected sample tubes are registered in the database using a QR code scanner and assigned to a 96-well sample plate in a layout that is kept throughout the assay.
- Interface RT-qPCR software (CFX Maestro)
- Export plate
Upon registration and primary processing of samples, the layout of the 96-well sample plate is exported for easy import into the RT-qPCR software (CFX Maestro). - Import analysis results
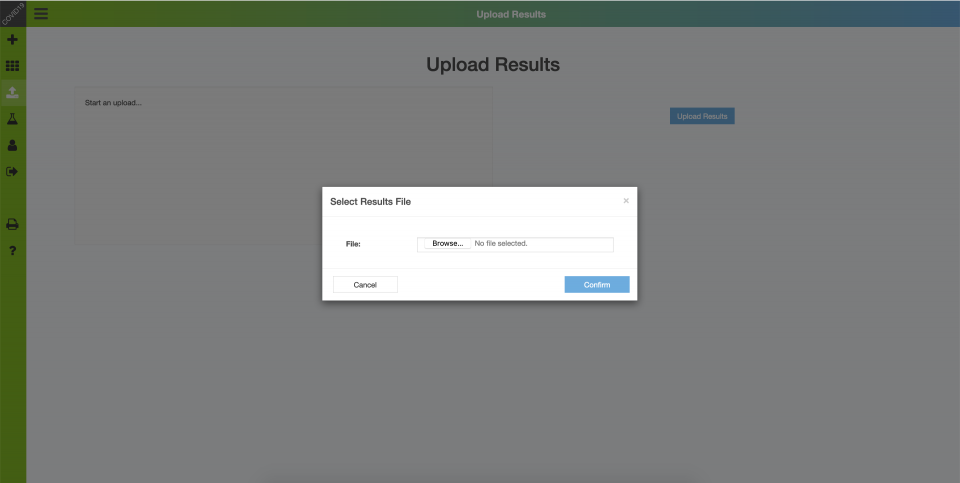
Upon completion of the RT-qPCR assay, results (i.e. Cq values and optional notes on individual results) are exported from the RT-qPCR software and imported into the database.
- Export plate
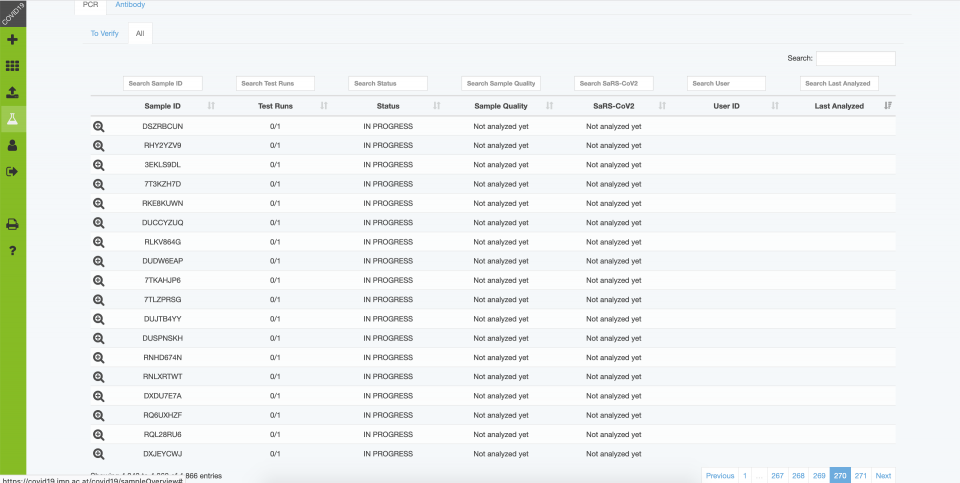
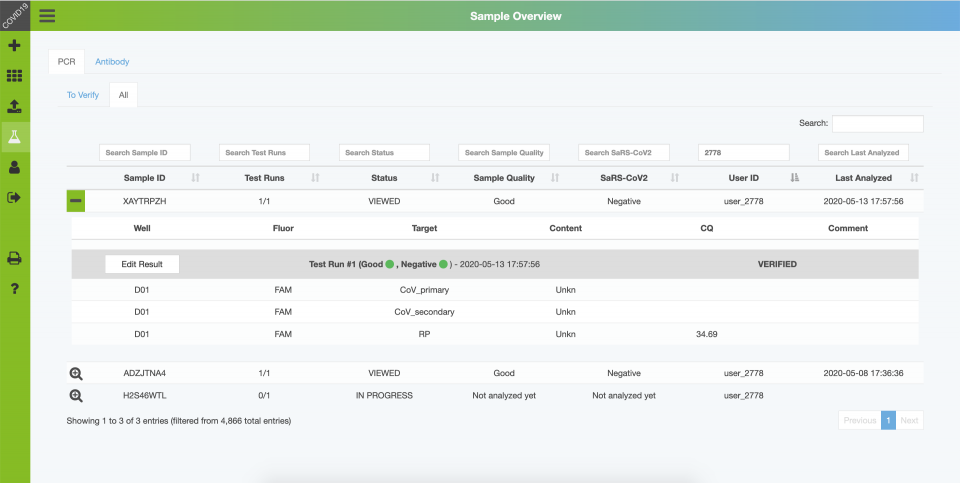
- Sample information and verification
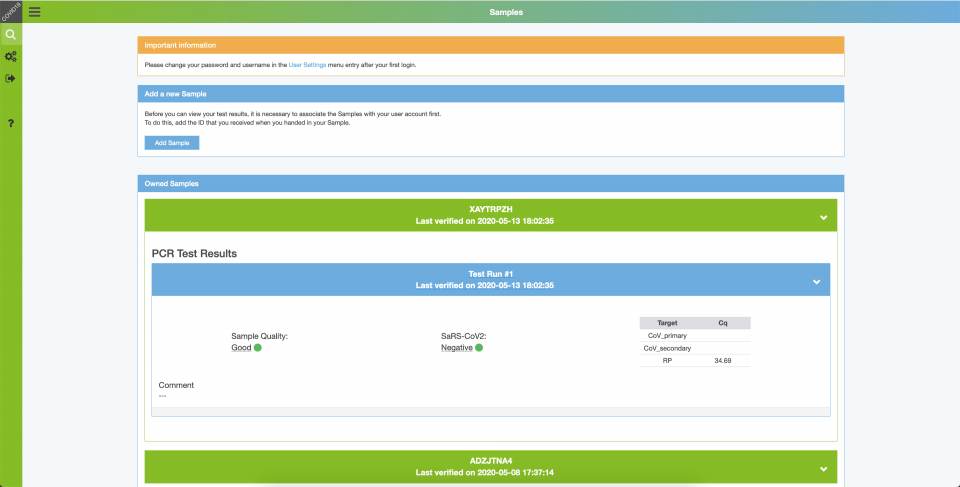
Results are then reviewed and interpreted by experienced personnel, and then released for distribution. Communicated results include the Cq values (e.g. N1, N2, RP), an interpretation of the test result and sample quality, and optional comments (e.g. to provide further interpretations and instructions in case of positive or ambiguous results).
- Patient frontend
Users obtain their test results by logging into the website and entering their 8-digit sample ID. Upon entry, the respective sampleID is permanently affiliated with the given user, so each patient can review all previous test results.
Flow charts
Login – Frontend/Backend
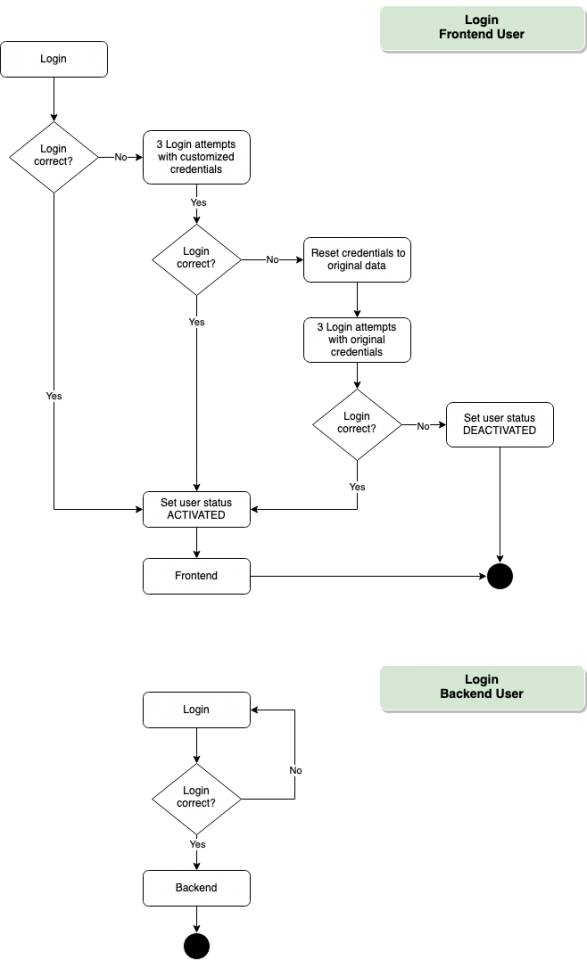
Frontend
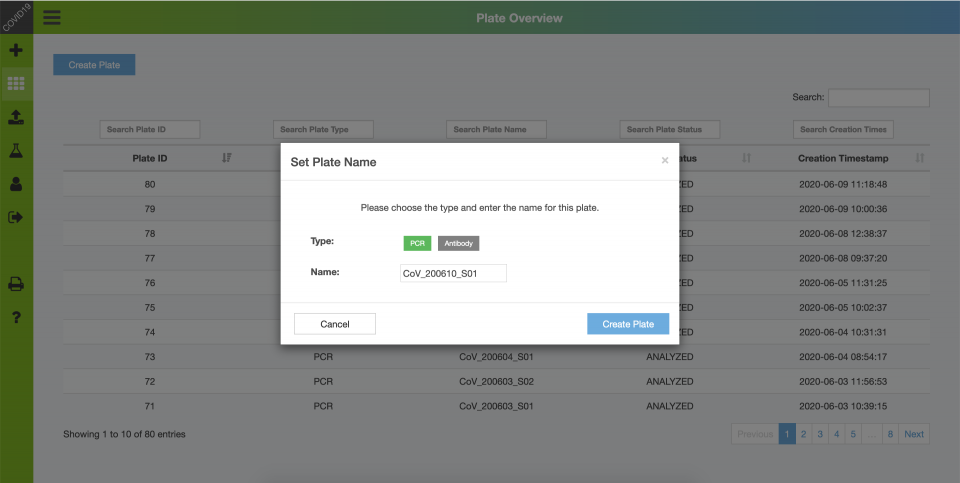
Backend – Create Samples
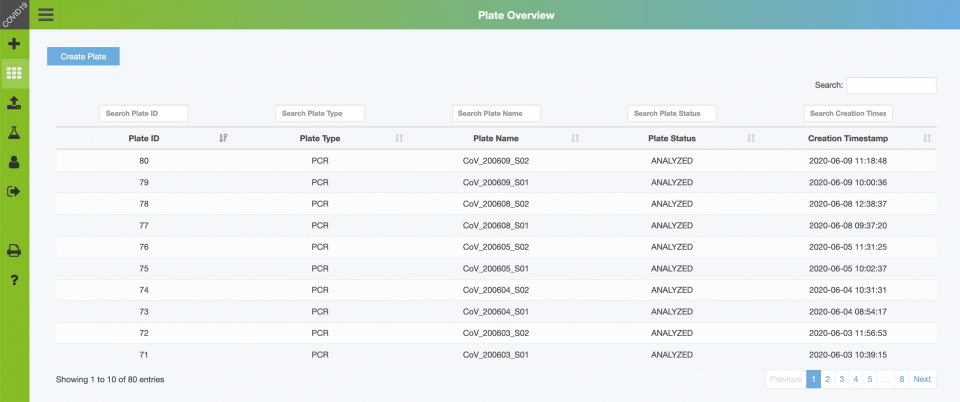
Backend – Plate Overview
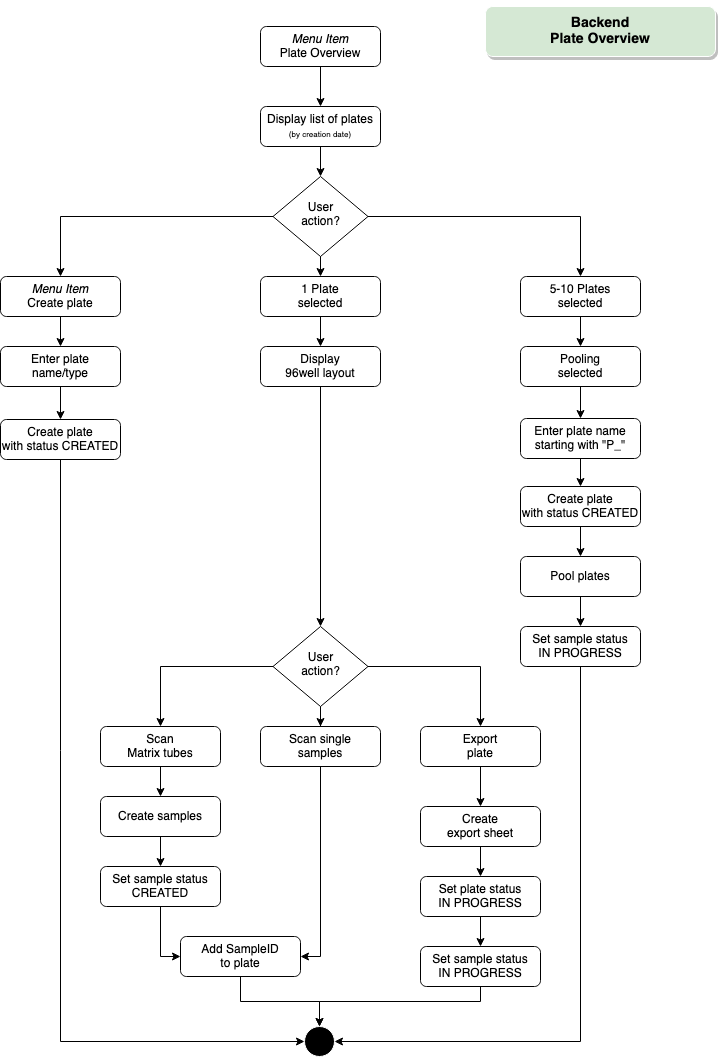
- Download here a demo file for the feature “Export plate” [csv]
Backend – Upload Results
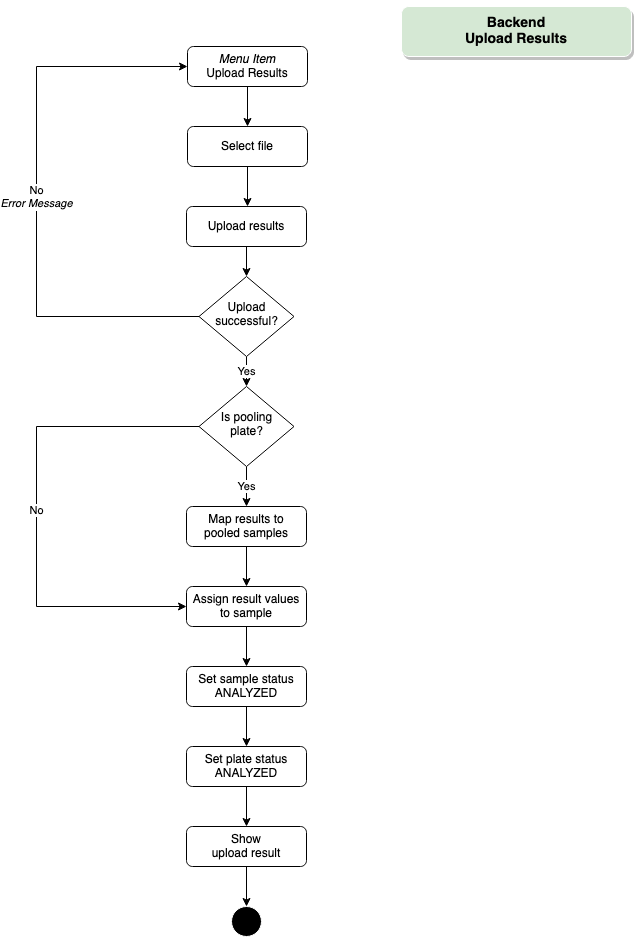
- Download here a demo files for the feature “Upload results”
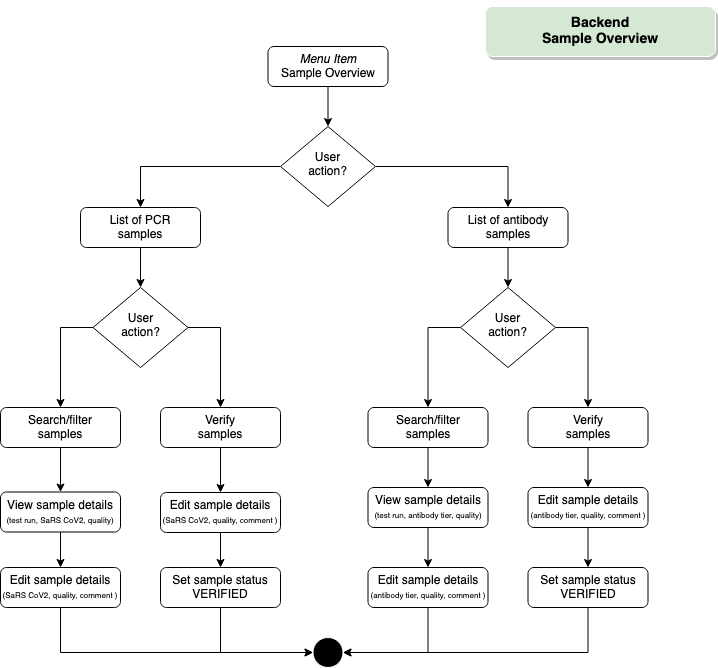
Backend – Sample Overview
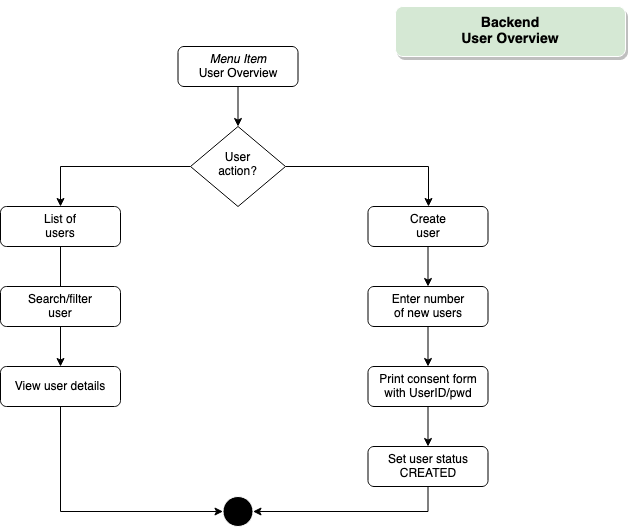
Backend – User Overview
Technical details
- Database management system
- PostgreSQL
- Application layer
- Java | Spring Boot framework
- Presentation layer
- Jakarta Server Pages
- Bootstrap | CSS framework
- JavaScript | Ajax with JQuery
- Development nodes
- MacBook Pro 2019 (2,4 GHz Quad-Core Intel Core i5, 16 GB RAM)
- MS Surface Pro 6 (Intel Core i5 2,4 GHz Quad-Core Intel Core i5, 16 GB RAM)
- Production server
- Intel(R) Xeon(R) Gold 6140 CPU @ 2.30GHz
- 2 processor with 2 cores
- 8 GB RAM
- Operating system: CentOS-7
- Software requirements:
- OpenJDK 18.9 (build 11.0.8+10-LTS)
- PostgreSQL 9.6.x
- Apache Maven 3.5.4
Resources
- Flow charts [zip]
- Plate export – demo file [csv]
- Result upload – demo file containing RP values [xlsx]
- Result upload – demo file containing primary/secondary values [xlsx]
Contact
Thomas Micheler
Strategic Information Management
VBCF – Vienna BioCenter Core Facilities GmbH
E-Mail: thomas.micheler@vbcf.ac.at
Phone: +43 664 80847 7130